- Learners can apply functions from the tidyr R Package to transform their data from a wide to a long format and vice versa.
- Learners can apply functions from the dplyr R Package to join multiple data sets.
- Learners can understand the relevance a data dictionary to describe variable names in a data set.
Pivoting & Joining & Metadata
ds4owd - data science for openwashdata
2023-12-12
Learning Objectives (for this week)
Homework module 6
Your turn: Discuss the reading
In discussion groups of 3, share your examples and discuss how the recommendations would improve your workflows.
In your homework you have read Wilson et al. (2017): “Good enough practices in scientific computing”.
You chose one of the recommended practices (Data Management, Software, Collaboration, Project organization, Keeping track of changes, Manuscripts), then:
You came up with a real-world example or scenario where the recommended practice could be applied in your research or academic work.
You are prepared to explain how the recommended practices would improve your workflows. This will be in the class setting as part of small discussion group (max 3 people).
10:00
Part 1: Pivoting data
Pivoting data
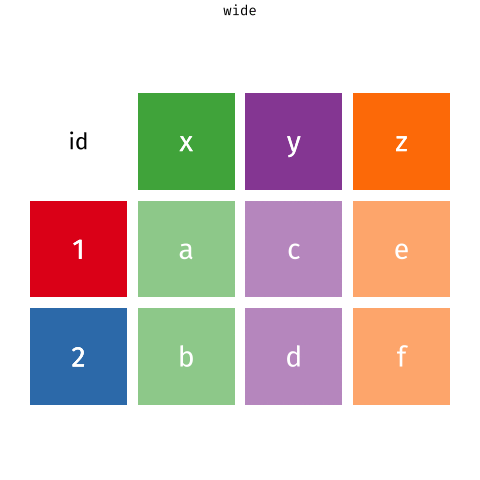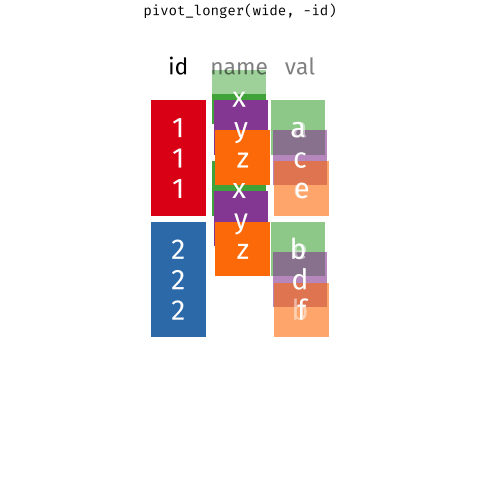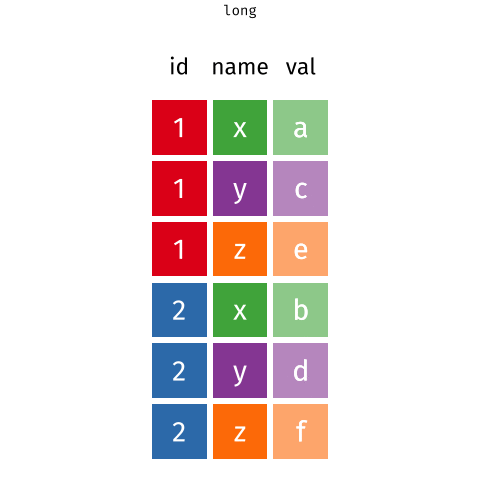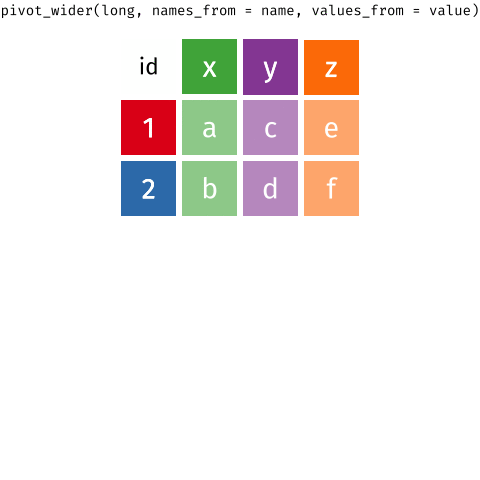
Waste characterisation data
| objid | location | pet | metal_alu | glass | paper | other | total |
|---|---|---|---|---|---|---|---|
| 900 | eth | 0.06 | 0.06 | 0.58 | 0.21 | 1.14 | 2.05 |
| 899 | eth | 0.14 | 0.01 | 0.18 | 0.28 | 3.04 | 3.64 |
| 921 | old_town | 0.00 | 0.00 | 0.00 | 0.41 | 1.57 | 1.99 |
| 916 | old_town | 0.17 | 0.04 | 0.80 | 0.55 | 0.62 | 2.19 |
| 900 | eth | 0.10 | 0.04 | 0.00 | 0.40 | 0.58 | 1.12 |
| 899 | eth | 0.08 | 0.03 | 0.00 | 0.05 | 0.34 | 0.50 |
| 921 | old_town | 0.08 | 0.03 | 0.30 | 0.40 | 1.52 | 2.33 |
| 916 | old_town | 0.11 | 0.04 | 0.92 | 1.01 | 1.99 | 4.07 |
How would you plot this?
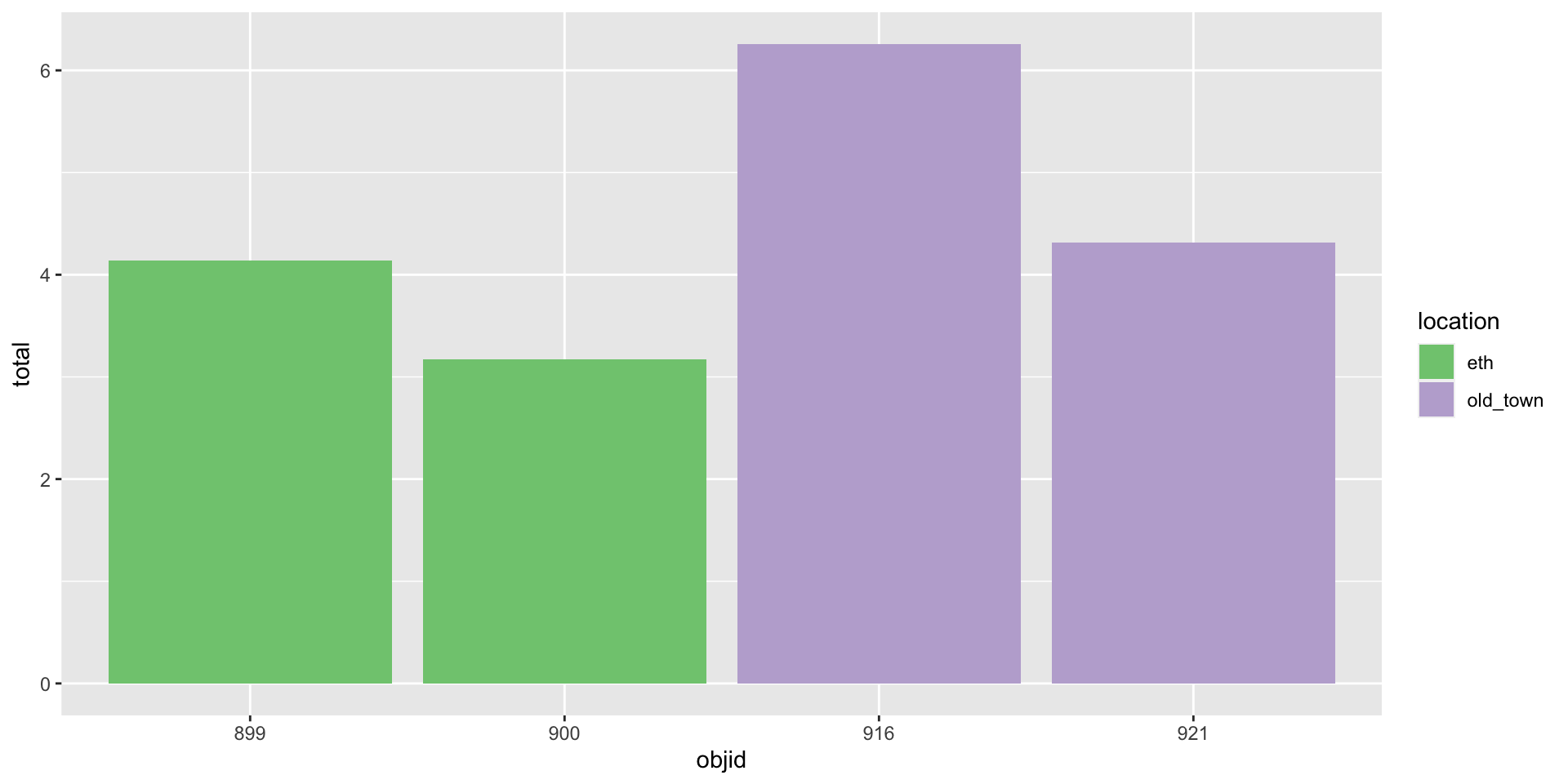
Three variables
| objid | location | total |
|---|---|---|
| 900 | eth | 2.05 |
| 899 | eth | 3.64 |
| 921 | old_town | 1.99 |
| 916 | old_town | 2.19 |
| 900 | eth | 1.12 |
| 899 | eth | 0.50 |
| 921 | old_town | 2.33 |
| 916 | old_town | 4.07 |
Three variables -> three aesthetics
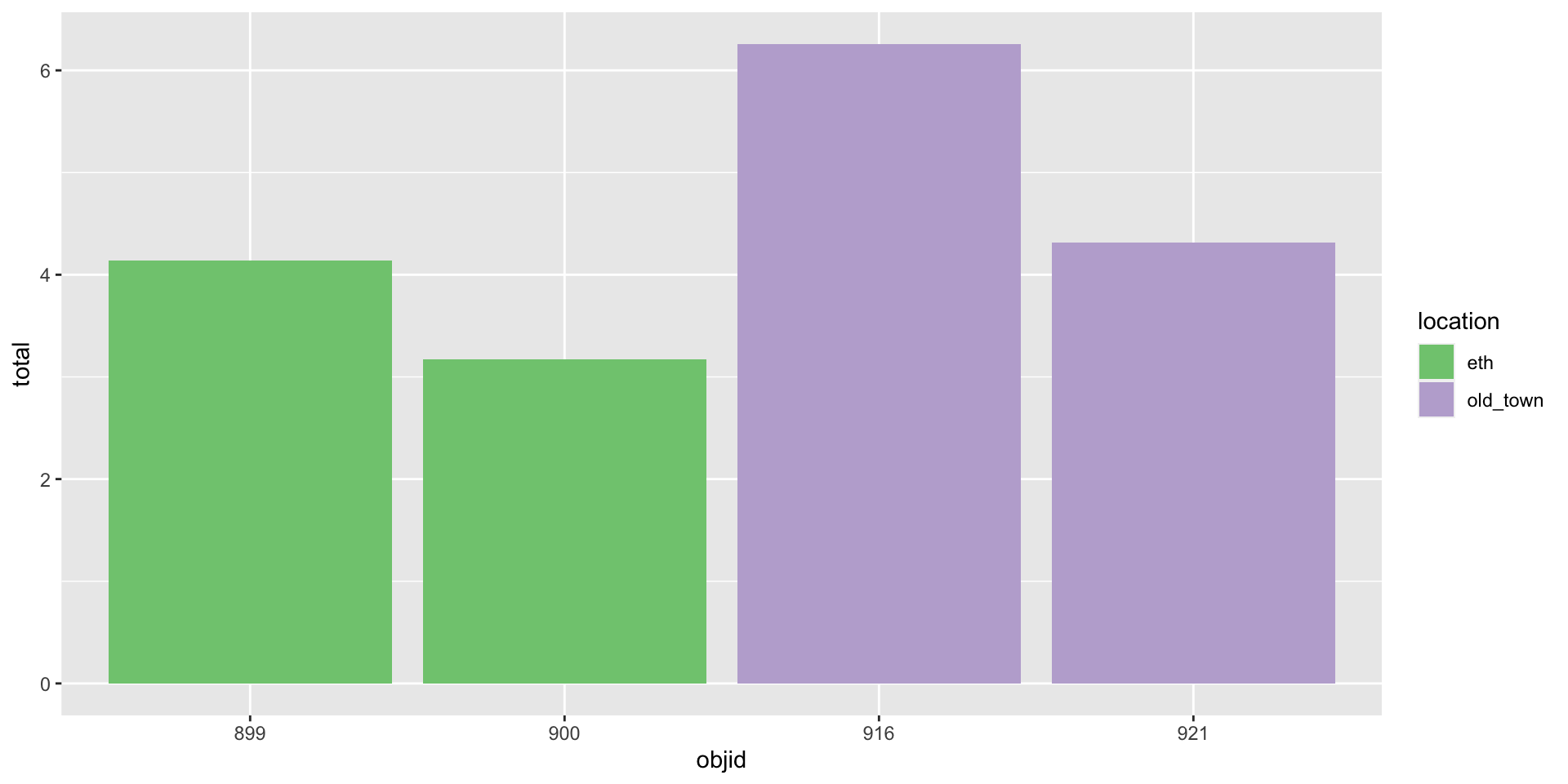
And how to plot this?
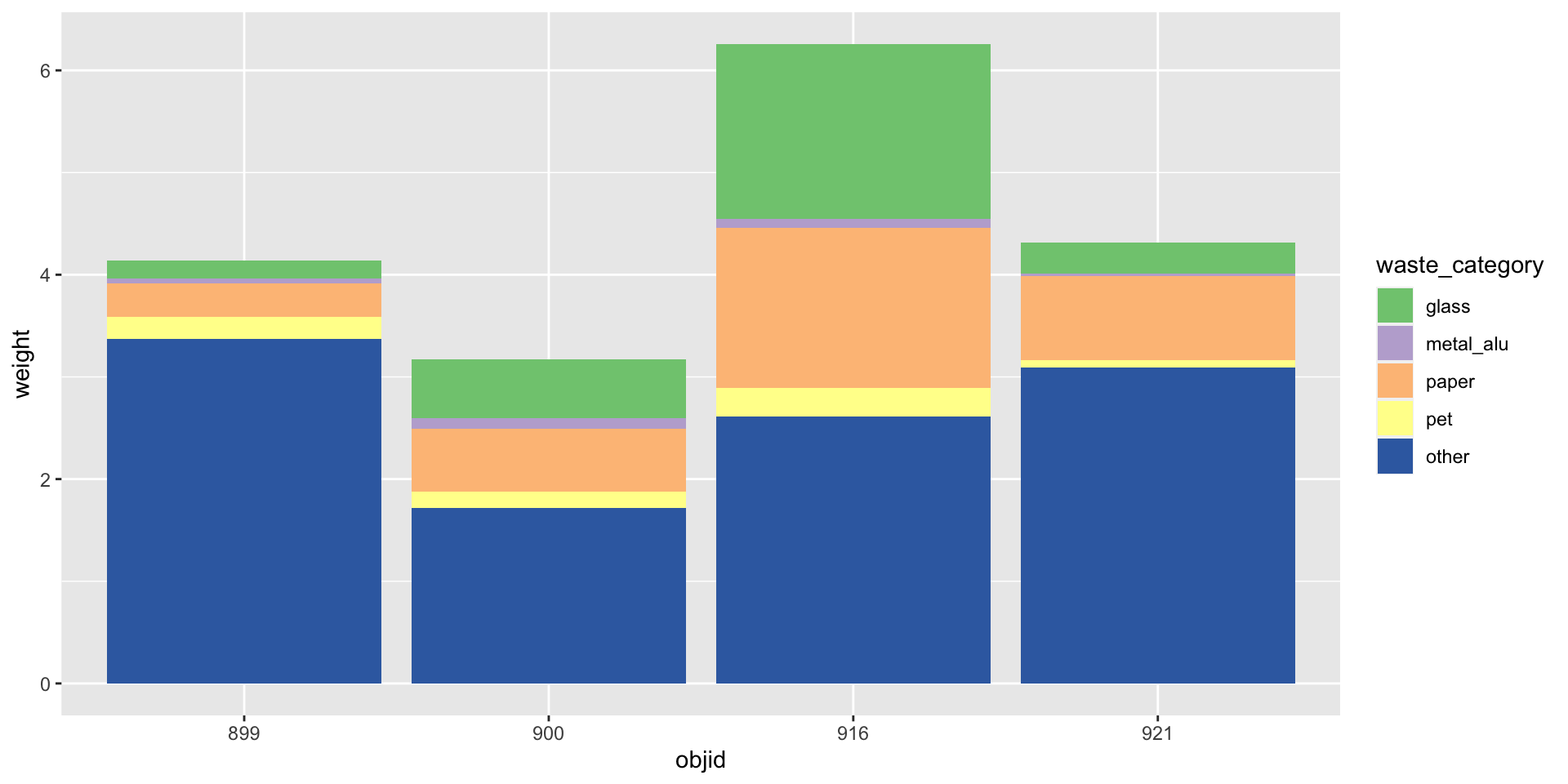
Reminder: Data (in wide format)
| objid | location | pet | metal_alu | glass | paper | other |
|---|---|---|---|---|---|---|
| 900 | eth | 0.06 | 0.06 | 0.58 | 0.21 | 1.14 |
| 899 | eth | 0.14 | 0.01 | 0.18 | 0.28 | 3.04 |
| 921 | old_town | 0.00 | 0.00 | 0.00 | 0.41 | 1.57 |
| 916 | old_town | 0.17 | 0.04 | 0.80 | 0.55 | 0.62 |
| 900 | eth | 0.10 | 0.04 | 0.00 | 0.40 | 0.58 |
| 899 | eth | 0.08 | 0.03 | 0.00 | 0.05 | 0.34 |
| 921 | old_town | 0.08 | 0.03 | 0.30 | 0.40 | 1.52 |
| 916 | old_town | 0.11 | 0.04 | 0.92 | 1.01 | 1.99 |
You need: A long format
| objid | location | waste_category | weight |
|---|---|---|---|
| 900 | eth | pet | 0.06 |
| 900 | eth | metal_alu | 0.06 |
| 900 | eth | glass | 0.58 |
| 900 | eth | paper | 0.21 |
| 900 | eth | other | 1.14 |
| 899 | eth | pet | 0.14 |
| 899 | eth | metal_alu | 0.01 |
| 899 | eth | glass | 0.18 |
| 899 | eth | paper | 0.28 |
| 899 | eth | other | 3.04 |
| 921 | old_town | pet | 0.00 |
| 921 | old_town | metal_alu | 0.00 |
| 921 | old_town | glass | 0.00 |
| 921 | old_town | paper | 0.41 |
| 921 | old_town | other | 1.57 |
| 916 | old_town | pet | 0.17 |
| 916 | old_town | metal_alu | 0.04 |
| 916 | old_town | glass | 0.80 |
| 916 | old_town | paper | 0.55 |
| 916 | old_town | other | 0.62 |
| 900 | eth | pet | 0.10 |
| 900 | eth | metal_alu | 0.04 |
| 900 | eth | glass | 0.00 |
| 900 | eth | paper | 0.40 |
| 900 | eth | other | 0.58 |
| 899 | eth | pet | 0.08 |
| 899 | eth | metal_alu | 0.03 |
| 899 | eth | glass | 0.00 |
| 899 | eth | paper | 0.05 |
| 899 | eth | other | 0.34 |
| 921 | old_town | pet | 0.08 |
| 921 | old_town | metal_alu | 0.03 |
| 921 | old_town | glass | 0.30 |
| 921 | old_town | paper | 0.40 |
| 921 | old_town | other | 1.52 |
| 916 | old_town | pet | 0.11 |
| 916 | old_town | metal_alu | 0.04 |
| 916 | old_town | glass | 0.92 |
| 916 | old_town | paper | 1.01 |
| 916 | old_town | other | 1.99 |
Three variables -> three aesthetics
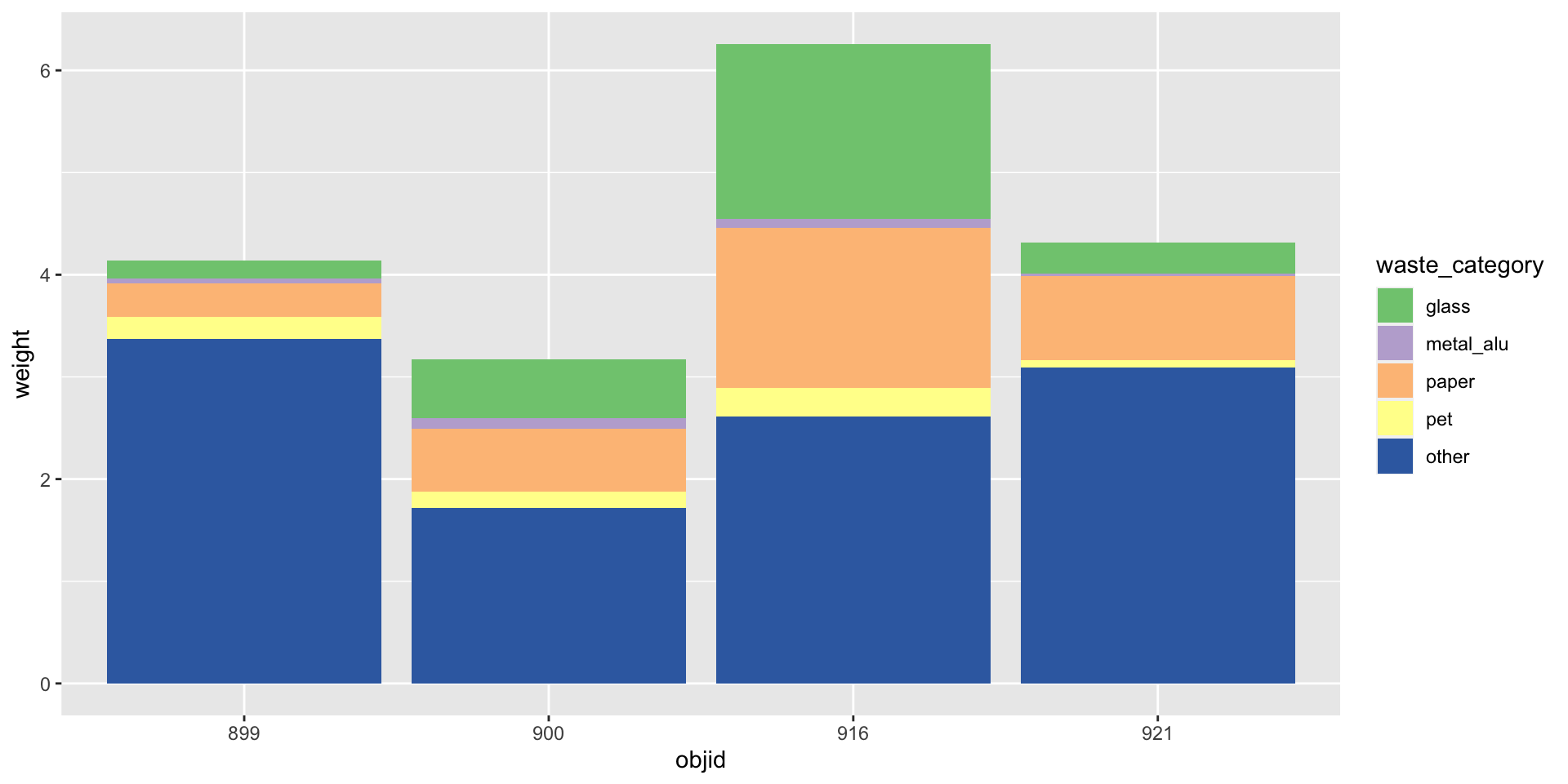
How to
| objid | location | pet | metal_alu | glass | paper | other |
|---|---|---|---|---|---|---|
| 900 | eth | 0.06 | 0.06 | 0.58 | 0.21 | 1.14 |
| 899 | eth | 0.14 | 0.01 | 0.18 | 0.28 | 3.04 |
| 921 | old_town | 0.00 | 0.00 | 0.00 | 0.41 | 1.57 |
| 916 | old_town | 0.17 | 0.04 | 0.80 | 0.55 | 0.62 |
| 900 | eth | 0.10 | 0.04 | 0.00 | 0.40 | 0.58 |
| 899 | eth | 0.08 | 0.03 | 0.00 | 0.05 | 0.34 |
| 921 | old_town | 0.08 | 0.03 | 0.30 | 0.40 | 1.52 |
| 916 | old_town | 0.11 | 0.04 | 0.92 | 1.01 | 1.99 |
How to
| objid | location | waste_category | weight |
|---|---|---|---|
| 900 | eth | pet | 0.06 |
| 900 | eth | metal_alu | 0.06 |
| 900 | eth | glass | 0.58 |
| 900 | eth | paper | 0.21 |
| 900 | eth | other | 1.14 |
| 899 | eth | pet | 0.14 |
| 899 | eth | metal_alu | 0.01 |
| 899 | eth | glass | 0.18 |
| 899 | eth | paper | 0.28 |
| 899 | eth | other | 3.04 |
| 921 | old_town | pet | 0.00 |
| 921 | old_town | metal_alu | 0.00 |
| 921 | old_town | glass | 0.00 |
| 921 | old_town | paper | 0.41 |
| 921 | old_town | other | 1.57 |
| 916 | old_town | pet | 0.17 |
| 916 | old_town | metal_alu | 0.04 |
| 916 | old_town | glass | 0.80 |
| 916 | old_town | paper | 0.55 |
| 916 | old_town | other | 0.62 |
| 900 | eth | pet | 0.10 |
| 900 | eth | metal_alu | 0.04 |
| 900 | eth | glass | 0.00 |
| 900 | eth | paper | 0.40 |
| 900 | eth | other | 0.58 |
| 899 | eth | pet | 0.08 |
| 899 | eth | metal_alu | 0.03 |
| 899 | eth | glass | 0.00 |
| 899 | eth | paper | 0.05 |
| 899 | eth | other | 0.34 |
| 921 | old_town | pet | 0.08 |
| 921 | old_town | metal_alu | 0.03 |
| 921 | old_town | glass | 0.30 |
| 921 | old_town | paper | 0.40 |
| 921 | old_town | other | 1.52 |
| 916 | old_town | pet | 0.11 |
| 916 | old_town | metal_alu | 0.04 |
| 916 | old_town | glass | 0.92 |
| 916 | old_town | paper | 1.01 |
| 916 | old_town | other | 1.99 |
Three variables -> three aesthetics
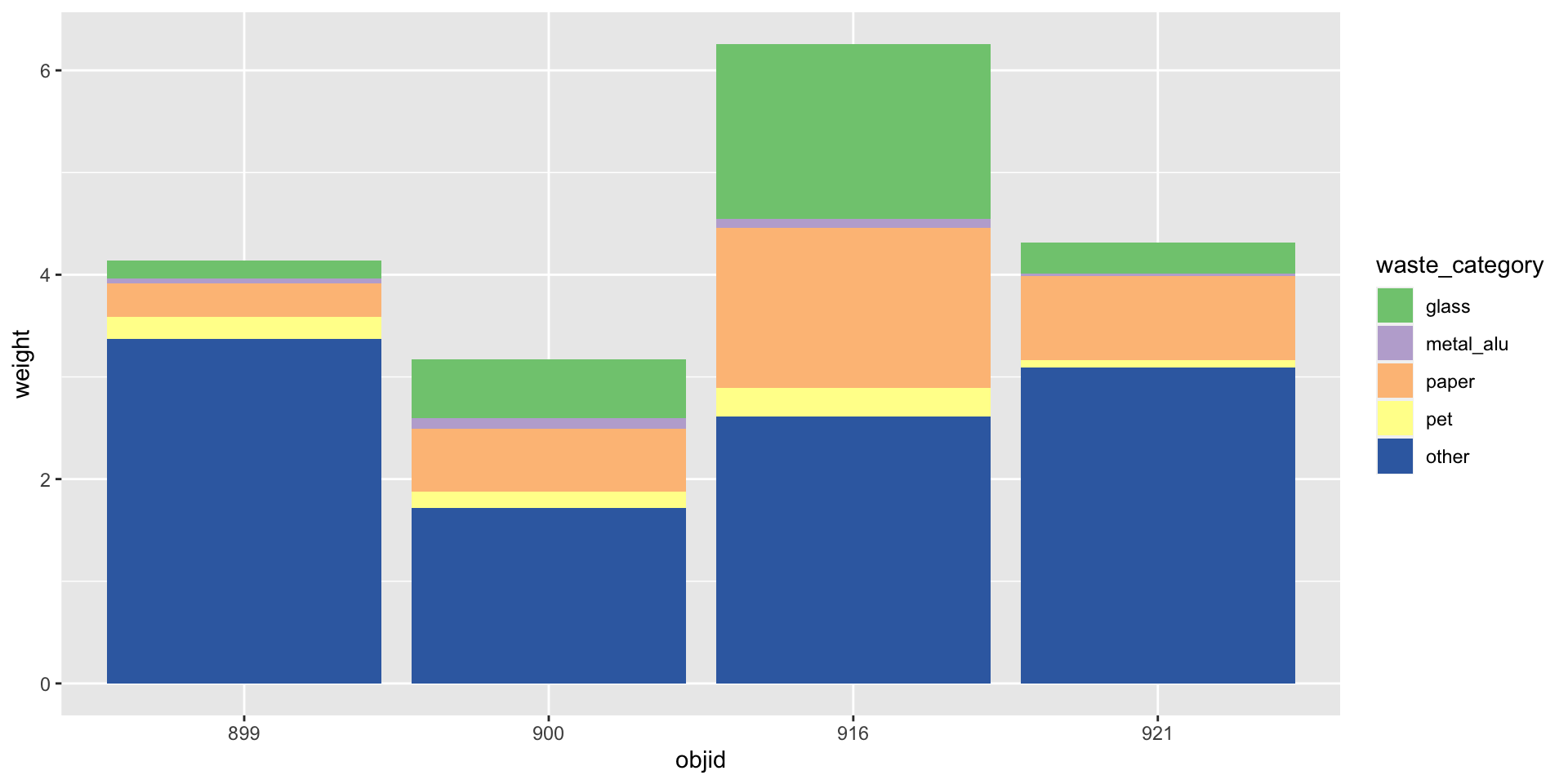
Your turn: md-07-exercises - pivoting data
- Open posit.cloud in your browser (use your bookmark).
- Open the ds4owd workspace for the course.
- In the File Manager in the bottom right window, locate the
md-07a-pivoting-your-turn.qmdfile and click on it to open it in the top left window. - Follow instructions in the file
30:00
Take a break
Please get up and move! Let your emails rest in peace.
10:00
Image generated with DALL-E 3 by OpenAI
Part 2: Joining data
We…
…have multiple data frames
…want to bring them together
Data: Women in science
Information on 10 women in science who changed the world
| name |
|---|
| Ada Lovelace |
| Marie Curie |
| Janaki Ammal |
| Chien-Shiung Wu |
| Katherine Johnson |
| Rosalind Franklin |
| Vera Rubin |
| Gladys West |
| Flossie Wong-Staal |
| Jennifer Doudna |
Inputs
| name | profession |
|---|---|
| Ada Lovelace | Mathematician |
| Marie Curie | Physicist and Chemist |
| Janaki Ammal | Botanist |
| Chien-Shiung Wu | Physicist |
| Katherine Johnson | Mathematician |
| Rosalind Franklin | Chemist |
| Vera Rubin | Astronomer |
| Gladys West | Mathematician |
| Flossie Wong-Staal | Virologist and Molecular Biologist |
| Jennifer Doudna | Biochemist |
| name | birth_year | death_year |
|---|---|---|
| Janaki Ammal | 1897 | 1984 |
| Chien-Shiung Wu | 1912 | 1997 |
| Katherine Johnson | 1918 | 2020 |
| Rosalind Franklin | 1920 | 1958 |
| Vera Rubin | 1928 | 2016 |
| Gladys West | 1930 | NA |
| Flossie Wong-Staal | 1947 | NA |
| Jennifer Doudna | 1964 | NA |
| name | known_for |
|---|---|
| Ada Lovelace | first computer algorithm |
| Marie Curie | theory of radioactivity, discovery of elements polonium and radium, first woman to win a Nobel Prize |
| Janaki Ammal | hybrid species, biodiversity protection |
| Chien-Shiung Wu | confim and refine theory of radioactive beta decy, Wu experiment overturning theory of parity |
| Katherine Johnson | calculations of orbital mechanics critical to sending the first Americans into space |
| Vera Rubin | existence of dark matter |
| Gladys West | mathematical modeling of the shape of the Earth which served as the foundation of GPS technology |
| Flossie Wong-Staal | first scientist to clone HIV and create a map of its genes which led to a test for the virus |
| Jennifer Doudna | one of the primary developers of CRISPR, a ground-breaking technology for editing genomes |
Desired output
| name | profession | birth_year | death_year | known_for |
|---|---|---|---|---|
| Ada Lovelace | Mathematician | NA | NA | first computer algorithm |
| Marie Curie | Physicist and Chemist | NA | NA | theory of radioactivity, discovery of elements polonium and radium, first woman to win a Nobel Prize |
| Janaki Ammal | Botanist | 1897 | 1984 | hybrid species, biodiversity protection |
| Chien-Shiung Wu | Physicist | 1912 | 1997 | confim and refine theory of radioactive beta decy, Wu experiment overturning theory of parity |
| Katherine Johnson | Mathematician | 1918 | 2020 | calculations of orbital mechanics critical to sending the first Americans into space |
| Rosalind Franklin | Chemist | 1920 | 1958 | NA |
| Vera Rubin | Astronomer | 1928 | 2016 | existence of dark matter |
| Gladys West | Mathematician | 1930 | NA | mathematical modeling of the shape of the Earth which served as the foundation of GPS technology |
| Flossie Wong-Staal | Virologist and Molecular Biologist | 1947 | NA | first scientist to clone HIV and create a map of its genes which led to a test for the virus |
| Jennifer Doudna | Biochemist | 1964 | NA | one of the primary developers of CRISPR, a ground-breaking technology for editing genomes |
Inputs, reminder
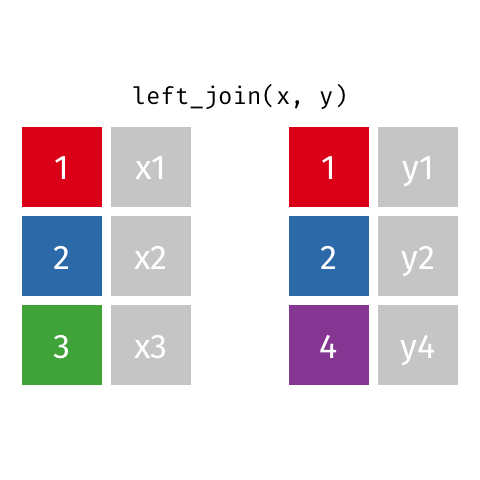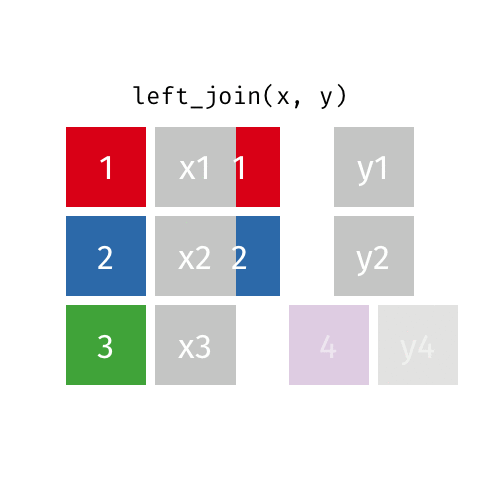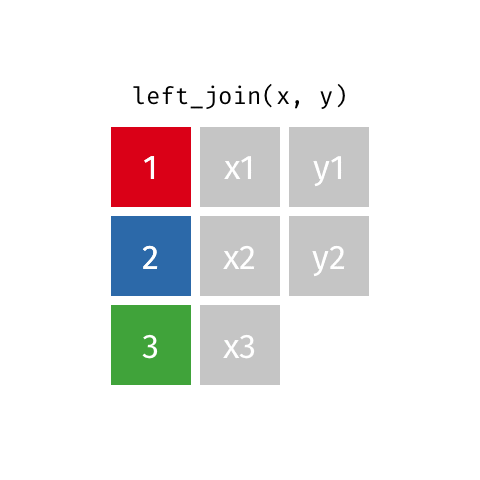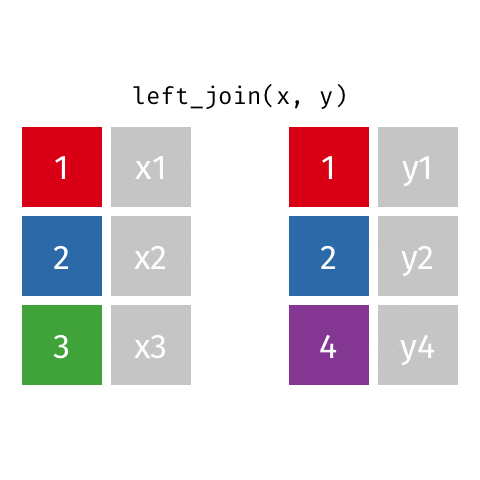
Joining data frames
left_join(): all rows from xright_join(): all rows from yfull_join(): all rows from both x and y- …
Setup
For the next few slides…
left_join()
left_join()
| name | profession | birth_year | death_year |
|---|---|---|---|
| Ada Lovelace | Mathematician | NA | NA |
| Marie Curie | Physicist and Chemist | NA | NA |
| Janaki Ammal | Botanist | 1897 | 1984 |
| Chien-Shiung Wu | Physicist | 1912 | 1997 |
| Katherine Johnson | Mathematician | 1918 | 2020 |
| Rosalind Franklin | Chemist | 1920 | 1958 |
| Vera Rubin | Astronomer | 1928 | 2016 |
| Gladys West | Mathematician | 1930 | NA |
| Flossie Wong-Staal | Virologist and Molecular Biologist | 1947 | NA |
| Jennifer Doudna | Biochemist | 1964 | NA |
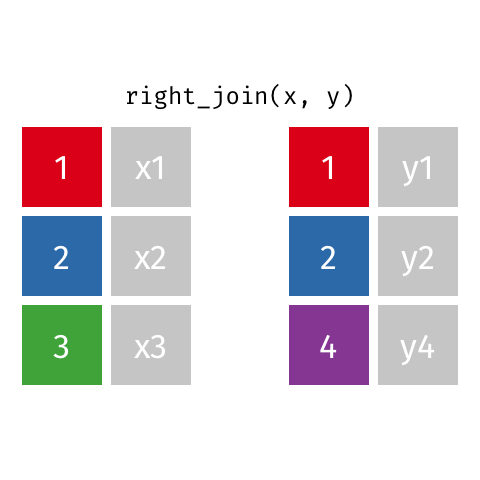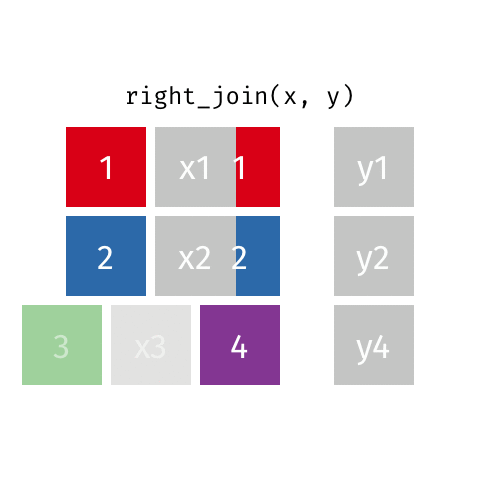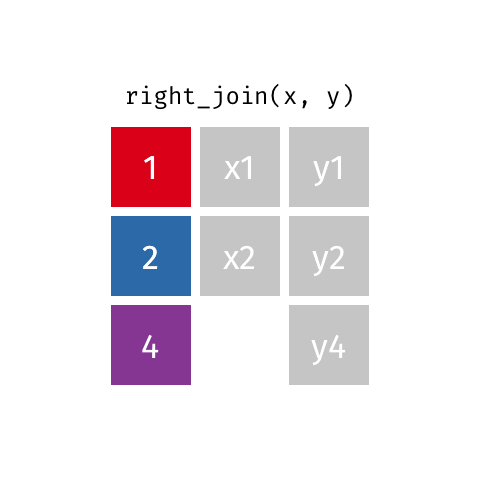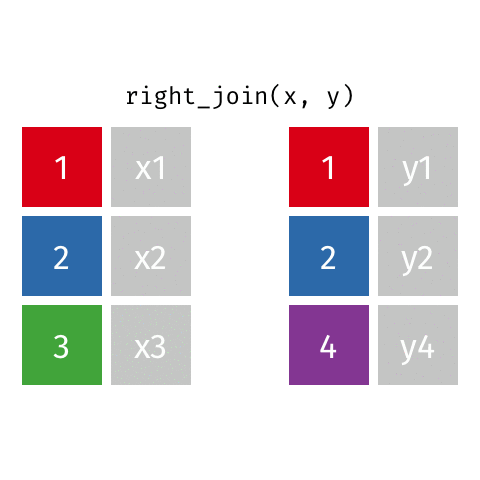
right_join()
right_join()
| name | profession | birth_year | death_year |
|---|---|---|---|
| Janaki Ammal | Botanist | 1897 | 1984 |
| Chien-Shiung Wu | Physicist | 1912 | 1997 |
| Katherine Johnson | Mathematician | 1918 | 2020 |
| Rosalind Franklin | Chemist | 1920 | 1958 |
| Vera Rubin | Astronomer | 1928 | 2016 |
| Gladys West | Mathematician | 1930 | NA |
| Flossie Wong-Staal | Virologist and Molecular Biologist | 1947 | NA |
| Jennifer Doudna | Biochemist | 1964 | NA |
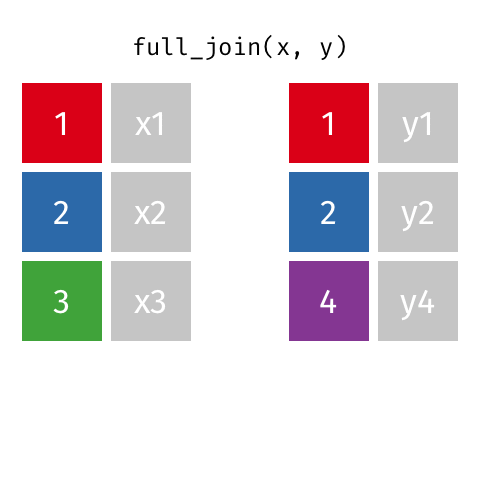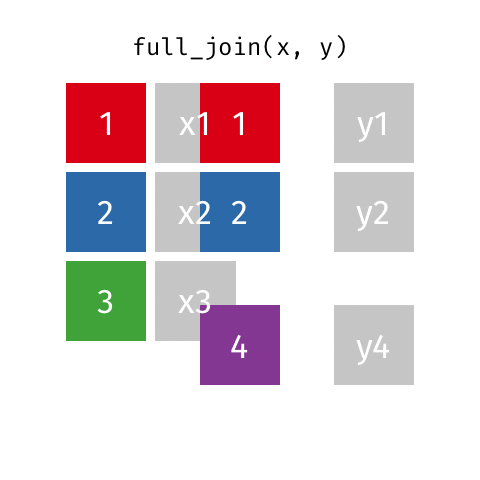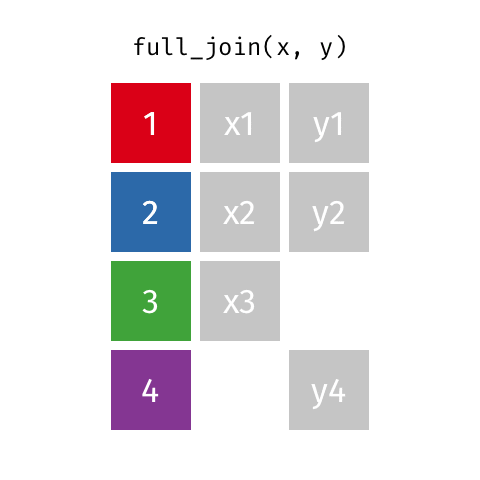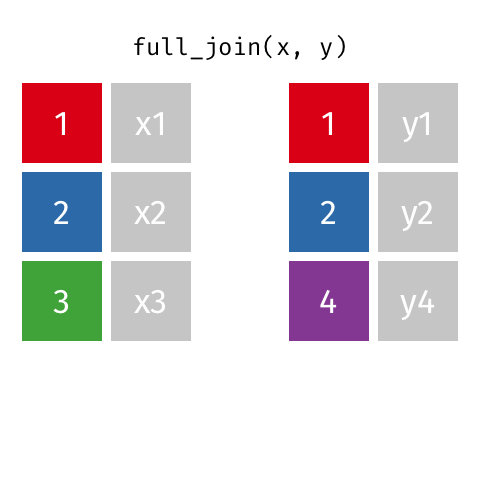
full_join()
full_join()
| name | birth_year | death_year | known_for |
|---|---|---|---|
| Janaki Ammal | 1897 | 1984 | hybrid species, biodiversity protection |
| Chien-Shiung Wu | 1912 | 1997 | confim and refine theory of radioactive beta decy, Wu experiment overturning theory of parity |
| Katherine Johnson | 1918 | 2020 | calculations of orbital mechanics critical to sending the first Americans into space |
| Rosalind Franklin | 1920 | 1958 | NA |
| Vera Rubin | 1928 | 2016 | existence of dark matter |
| Gladys West | 1930 | NA | mathematical modeling of the shape of the Earth which served as the foundation of GPS technology |
| Flossie Wong-Staal | 1947 | NA | first scientist to clone HIV and create a map of its genes which led to a test for the virus |
| Jennifer Doudna | 1964 | NA | one of the primary developers of CRISPR, a ground-breaking technology for editing genomes |
| Ada Lovelace | NA | NA | first computer algorithm |
| Marie Curie | NA | NA | theory of radioactivity, discovery of elements polonium and radium, first woman to win a Nobel Prize |
Putting it altogether
| name | profession | birth_year | death_year | known_for |
|---|---|---|---|---|
| Ada Lovelace | Mathematician | NA | NA | first computer algorithm |
| Marie Curie | Physicist and Chemist | NA | NA | theory of radioactivity, discovery of elements polonium and radium, first woman to win a Nobel Prize |
| Janaki Ammal | Botanist | 1897 | 1984 | hybrid species, biodiversity protection |
| Chien-Shiung Wu | Physicist | 1912 | 1997 | confim and refine theory of radioactive beta decy, Wu experiment overturning theory of parity |
| Katherine Johnson | Mathematician | 1918 | 2020 | calculations of orbital mechanics critical to sending the first Americans into space |
| Rosalind Franklin | Chemist | 1920 | 1958 | NA |
| Vera Rubin | Astronomer | 1928 | 2016 | existence of dark matter |
| Gladys West | Mathematician | 1930 | NA | mathematical modeling of the shape of the Earth which served as the foundation of GPS technology |
| Flossie Wong-Staal | Virologist and Molecular Biologist | 1947 | NA | first scientist to clone HIV and create a map of its genes which led to a test for the virus |
| Jennifer Doudna | Biochemist | 1964 | NA | one of the primary developers of CRISPR, a ground-breaking technology for editing genomes |
Part 3: Metadata
Metadata: data about data
WHAT!?
Faecal sludge samples
Imagine:
- you are new to WASH research and you have never heard of feacal sludge management.
- you are interested in learning more about the topic and you want to find some data to play with.
- you find a publication with a dataset on faecal sludge characteristics.
Faecal sludge samples
You download the XLSX file that contains the data and you open it in Excel. You see the following:
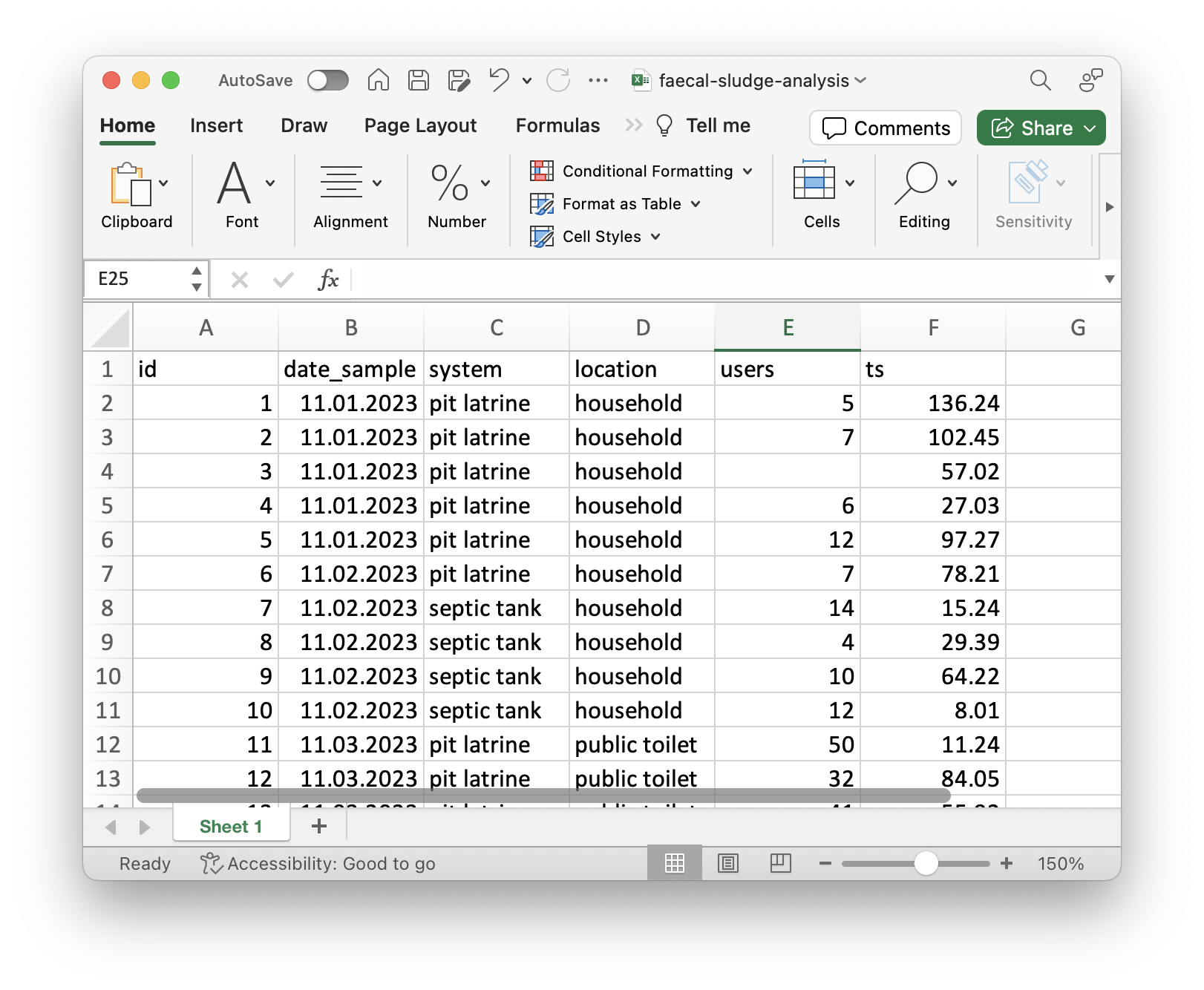
Faecal sludge samples
Open questions:
- What unit does
usersrefer to? - What does
tsstand for? - The
dateof what? - Where was this data collected?
- Which method was used to collect the samples?

Questions that only the original author may have the answers to.
You as an author
have the chance to document your data properly once to make it easier for everyone else to know what it contains.
Documentation
Goes into a separate README file
- General information (authors, title, date, geographic location, etc.)
- Sharing / access information (license, links to publications, citation)
- Methodological information (sampling, analysis, etc.)
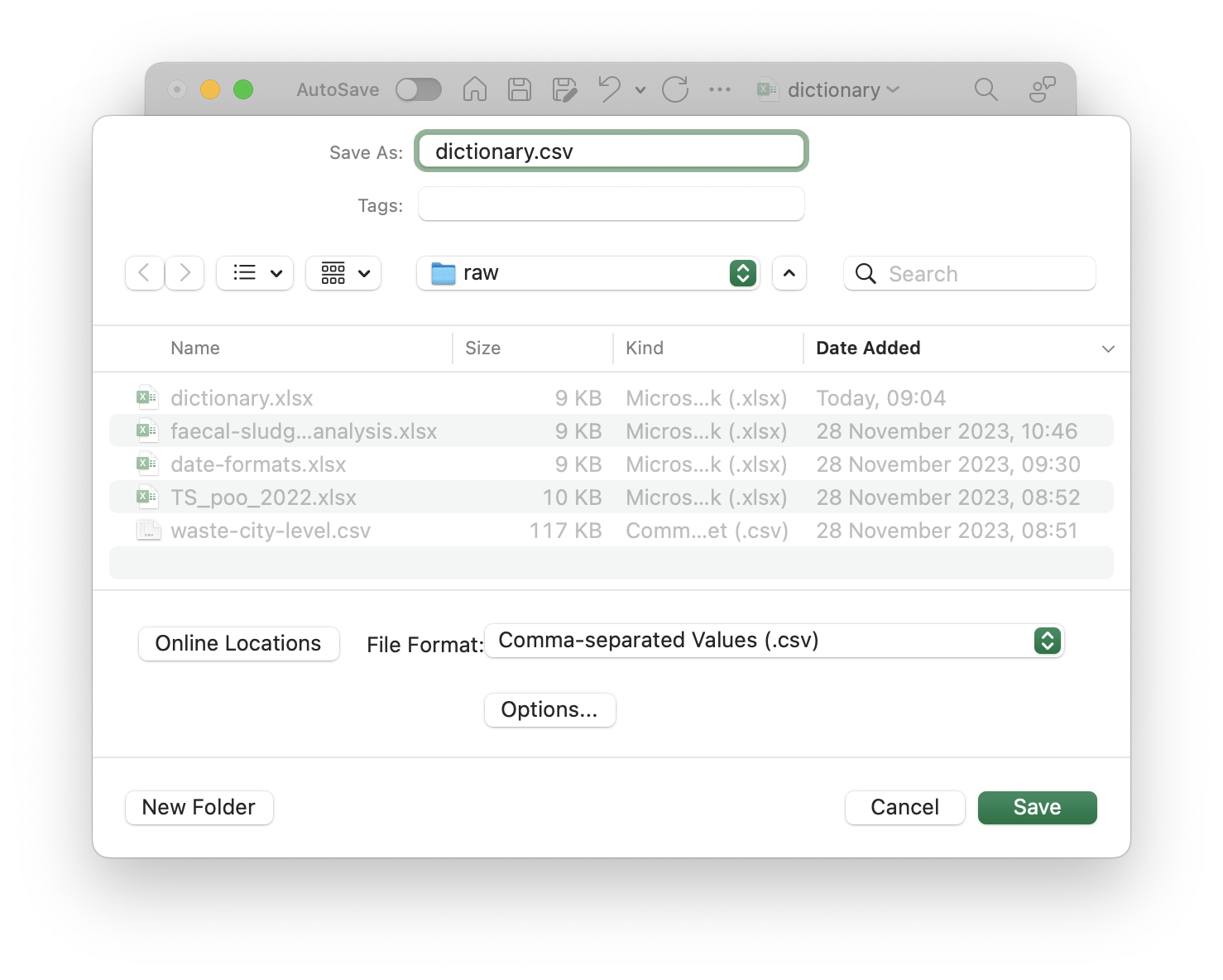
Data dictionary
Goes into a separate file (dictionary.csv).
Minimum required information
- Variable name
- Variable description
Data dictionary for faecal sludge samples
- Edit in spreadsheet software (e.g. MS Excel)

Data dictionary for faecal sludge samples
- Save as CSV file
Directory tree of a project
Capstone project of Rainbow Train: https://github.com/ds4owd-001/project-rainbow-train
.
├── R
│ └── 01-data-preparation.R
├── data
│ ├── processed
│ │ ├── README.md
│ │ ├── dictionary.csv
│ │ └── faecal-sludge-analysis.csv
│ └── raw
│ └── Faecal sludge Analysis_05112023.xlsx
├── docs
│ ├── index.html
│ ├── index.qmd
│ └── index_files
│ └── libs
└── project.RprojDirectory tree of a project
Rfolder: R scripts for data cleaningdatafolder: raw and processed datadocsfolder: the actual report that imports the processed dataproject.Rproj: RStudio project file
Inside the data folder
raw: data as it was downloaded / as you received it (e.g. Excel file)processed: data that is ready to be used in the report
Inside the processed folder
README.md: general information about the datadictionary.csv: data dictionaryfaecal-sludge-analysis.csv: cleaned data for whichdictionary.csvapplies
My turn: A tour of Rainbow Train’s project
Sit back and enjoy!
10:00
Homework assignments module 7
Module 7 documentation
Homework due date
- Homework assignment due: Monday, December 18th
- Feedback until: Thursday, December 21st
Wrap-up
Final student hours of 2023
- Thursday, December 14th at 2 pm CET
First student hours of 2024
- Thursday, January 11th at 2 pm CET
Christmas break
- Lars will be on vacation from December 22nd until January 15th
- Mian and Sophia will be on vacation from December 22nd until January 8th

Image generated with DALL-E 3 by OpenAI
First lecture of 2024
Tuesday, January 16th at 2 pm CET

Image generated with DALL-E 3 by OpenAI
Course calendar
| date | week | topic | module |
|---|---|---|---|
| 31 October 2023 | 1 | Welcome & get ready for the course | module 1 |
| 07 November 2023 | 2 | Data science lifecycle & Exploratory data analysis using visualization | module 2 |
| 14 November 2023 | 3 | Data transformation with dplyr | module 3 |
| 21 November 2023 | 4 | Data import & Data organization in spreadsheets | module 4 |
| 28 November 2023 | 5 | Conditions & Dates & Tables | module 5 |
| 05 December 2023 | 6 | Data types & Vectors & For Loops | module 6 |
| 12 December 2023 | 7 | Pivoting & joining data | module 7 |
| 19 December 2023 | 8 | Break | NA |
| 26 December 2023 | 9 | Break | NA |
| 02 January 2024 | 10 | Break | NA |
| 09 January 2024 | 11 | Work on Capstone project | NA |
| 16 January 2024 | 12 | Writing functions & Creating a website with Quarto and GitHub pages | module 8 |
| 23 January 2024 | 13 | openwashdata webinar: a data sharing workflow that may please the publishers | NA |
| 30 January 2024 | 14 | Using AI for software development in R | module 9 |
| 06 February 2024 | 15 | Work on Capstone project | NA |
| 13 February 2024 | 16 | Final submission date of Capstone project | NA |
| 20 February 2024 | 17 | Graduation party of openwashdata academy | module 10 |
Thanks! 🌻
Slides created via revealjs and Quarto: https://quarto.org/docs/presentations/revealjs/ Access slides as PDF on GitHub
References
All material is licensed under Creative Commons Attribution Share Alike 4.0 International.